Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation[C]//International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241.
该篇论文
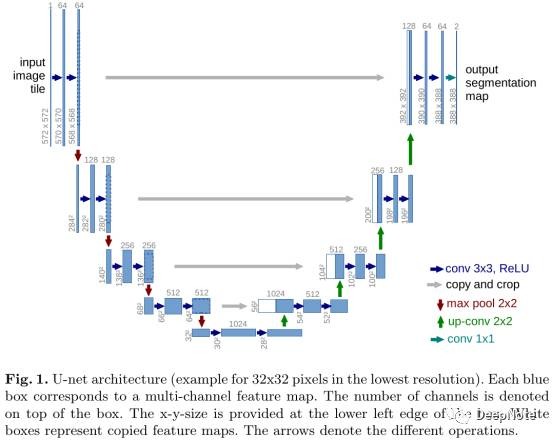
- 在FCN基础上提出U-Net结构 (Figure 1).
- 提出医疗影像data augmentation.
结合两者能够trained end-to-end from very few images and outperforms sliding-window CNN.
Localization: a class label is supposed to be assigned to each pixel.
1. Sliding-window drawbacks
- Network需要对每个patch单独处理,重叠的patch产生大量冗余,因此非常慢。
- Tradeoff between localization accuracy and the use of context. Large patches需要更多pooling层,导致localization accuracy下降,而small patches allow network see only little context.
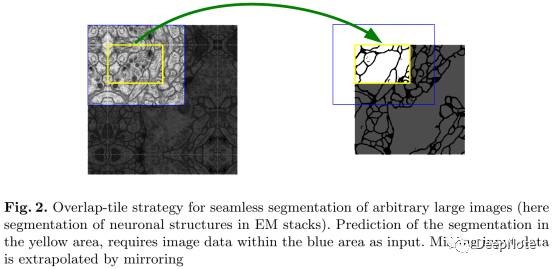
2. Overlap-tile Strategy (Figure 2)
- 该策略支持任意大小图片的无缝分割(seamless segmentation),蓝色区域为输入patch,黄色区域为输出patch(输入图片进行镜像处理).
3. Data Augmentation
- Shift and rotation invariance.
- Deformations and gray value invariance.
- Elastic deformation非常重要,能够有效模拟组织(tissue)最常见的形变方式。
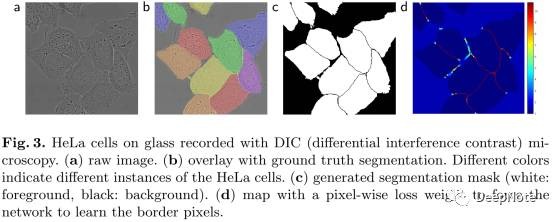
4. Touching Object Challenge (Figure 3)
- 分离同种类型接触的细胞。
- Propose the use of a weighted loss, where the separating background labels between touching cells obtain a large weight in the loss function.
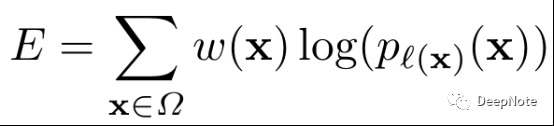
- 预先计算ground-truth的weight map, to force the network to learn the small separation borders that we introduce between touching cells.
- d1,d2: the distance to the border of the nearest and second nearest cell.
- wc: balance the class frequencies.
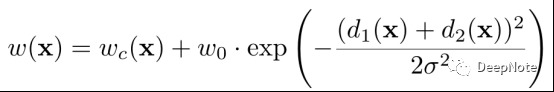
5. Initialization
- Ideally the initial weights should be adapted such that each feature map in the network has approximately unit variance.
- 论文采用Gaussian 方差srqt(2/$N$), $N$为输入Node数。例如3x3 Conv层64 kernels, 则$N$ = 9 * 64 = 576.
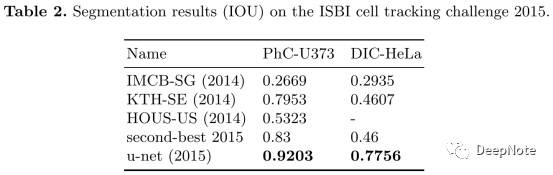
6. Experiment Results